Replication fork recovery after stalling is critical for cell survival and genome stability. My lab is interested in the mechanisms of replication restart in eukaryotes, how replication and recombination are coordinated and the consequences of mis-regulation for cancer progression. We use fission yeast as a model system to study how recombination is regulated at stalled forks to balance the need to restart replication with increased genome instability. Current projects focus on the roles of recombination regulators such as the Smc5/Smc6 complex in replication fork stability. SMC complexes are essential complexes involved in chromosome structure and segregation.
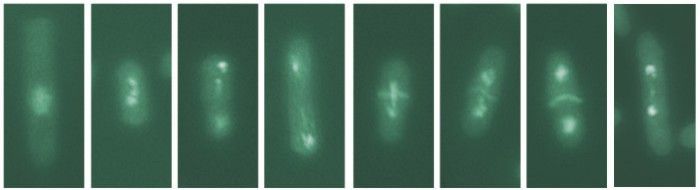
We use a combination of genome-wide and site-specific approaches to study the coordination of replication with recombination. In collaboration with Tony Carr we have developed an inducible system to study replication stalling within a short palindrome, where the DNA sequence reads the same backwards as forwards. In this system 25% of cells undergo chromosomal rearrangement each S phase resulting in cell death due to chromosome mis-segregation, enabling us to characterize the DNA intermediates formed and the pathways involved.