STRUCTURAL BIOLOGY OF CHROMATIN REMODELLING AND TRANSCRIPTIONAL REGULATION
We have previously dissected the molecular mechanism of action of a class of RNA packaging ATPases which act as molecular motors, providing a structural explanation for how the chemical energy derived from the hydrolysis of ATP might be coupled to produce mechanical action. We want to understand at a molecular level how transcription factors and chromatin remodelling ATPases work, alone or in complex, to define an active or repressed gene transcription state. To obtain insights into these cellular processes, we use X-ray crystallography in combination with Cryo-EM, NMR, SAXS and a range of biochemical and biophysical techniques.
Recently, we were able to dissect the roles of the chromatin targeting and motor domains in human chromatin remodelling factor CHD4 and demonstrate that their activities are allosterically regulated. Since this combination of domains is a unifying feature of all chromatin remodellers, we are now verifying whether the proposed mechanism is a general paradigm for this important class of molecules.]
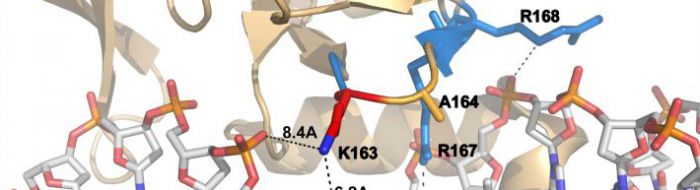
Finally, we are studying a multi-protein transcription factor complex, containing proteins SCL, LMO2 and GATA1, involved in erythroid cell lineage determination. We seek to understand the nature of the specific protein-protein protein-DNA interactions required in haemopoiesis, and also to obtain insights into the role of this complex in leukaemogenesis which may contribute to the design of novel targeted therapies. We have recently solved the structure the crystal structure of SCL:E47:LMO2:LDB1LID bound to DNA. Dissection at atomic level of the protein/protein and protein/DNA interactions of this complex combined with mammalian two-hybrid binding studies and in vivo gain of function studies in zebrafish embryos, reveal complex synergies between components of the complex their co-factors and DNA targets. This study deepens our understanding of how higher-order, hematopoietic-specific complexes form and how tissue-specific gene expression programs might be regulated. We are now expanding the structural analysis to include more protein partners including other transcription factors (such as GATA-1), co-activators, co-repressor and chromatin remodelling proteins.